From Jackknife to A/B Testing
Posted on 2019-12-23 22:28:32 +0900 in Data Science
Background
In A/B Testing, there is a group of data and , the metrics we interested in are the difference between these two groups and related confidence.
A Quick Introduction to Jackknife
Jackknife is a method of resample, which tries to estimate the bias and variability of an estimator by using values of on subsamples from .
The pseudovalue of is , where means the sample with value deleted from the sample.
Treat the pseudovalue as if they were independent random variables with mean , then the confidence interval could be obtained using Central Limit Theorem. Specifically, let
and
be the mean and sample variance of the pseudovalues. The jackknife 95% confidence interval is
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import math
sample_count = 500000
K = 5000
mu, sigma = 500, 100 # mean and standard deviation
def jackknife_sder(s):
df = pd.DataFrame(data = s, columns=['data'])
groups = int(sample_count / K )
def label_race (row):
return int(row.name) % groups
df['group'] = df.apply(lambda row: label_race(row), axis=1)
average = df['data'].mean()
total_sum = s.sum()
left_k_groups = [(total_sum - (df['data'][df['group'] == x]).sum()) / (sample_count - K) for x in range(groups)]
a = groups * average
b = [v * (groups - 1) for v in left_k_groups]
ps = a - b
mean, var = ps.mean(), (ps.var(ddof = 1.0) / groups) ** 0.5
return mean, var
data = [np.random.normal(mu, sigma, sample_count) for i in range(100)]
s_vars = [jackknife_sder(d) for d in data]
s_mean, s_var = mu, sigma / (sample_count ** 0.5)
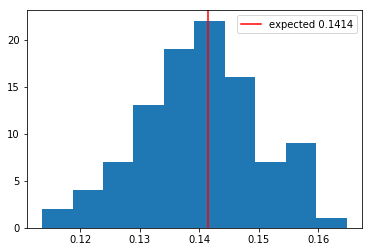
plt.hist(s_vars, bins=10)
plt.axvline(x=s_var, color='r', label=f'expected {s_var:.4f}')
plt.legend()
plt.show()
confidence = 1.96
l, r = (s_mean - confidence * s_vars, s_mean + confidence * s_vars)
print(f'left: {l:.4f}, right: {r:.4f}')
left: 499.7084, right: 500.2740